6 Where To Publish Data
6.1 Searching for Research data repositories
re3data is a global registry of research data repositories with over 3,000 entries where you can search for an appropriate place to deposit your data (Pampel et al. 2023 [cito:citesAsAuthority]). So if you don’t know of a suitable repository then searching for one that is a good fit for your data in re3data is a good place to start.
FAIRsharing.org is a curated resource of educational material on databases, standards, and policies for data sharing.
6.2 Selected Public Data Repositories & Data Sharing Platforms
6.2.1 Sequencing Data
6.2.1.1 GEO (Gene Expression Omnibus)
- Accepts data from methods which measure some property of genomic features e.g. expression micro-arrays, RNA-seq, ChIP-seq, ATAC-seq but not genomic sequence data.
- GEO submission
GEO has quite a flexible model for metadata you can (and should) include a good deal of additional data along with any sequencing data that you deposit here. They explicitly provide for secondary data (i.e. data derived from your sequencing data) to be included. It is also a good idea, wherever possible, to include the process (or appropriate links to) the process by which you got from the data/metadata to the secondary results. For example in previous submissions to GEO of RNA-seq data that I processed with a standard nf-core I have included the count matrices and some other pipeline outputs as secondary results. I also included command run to kick off the pipeline that generated these results, the design matrix input file needed in addition to the sequencing files and the version of the pipeline that I used. This way someone downloading my data could re-capitulate my results exactly by re-running that same version of the pipeline on my raw data. This also means anyone wanting to use my results can interrogate the code in the nf-core pipeline to see exactly how the analysis was performed.
6.2.1.2 SRA (Sequence Read Archive)
- Store the raw sequencing data underlying GEO, and other data including genomic sequencing data
6.2.1.3 HCA (Human Cell Atlas) data portal
The Human Cell Atlas project has a data portal
Much of HDBI’s data is likely to meet the data suitability criteria for the HCA data portal which are summarized in the image below.
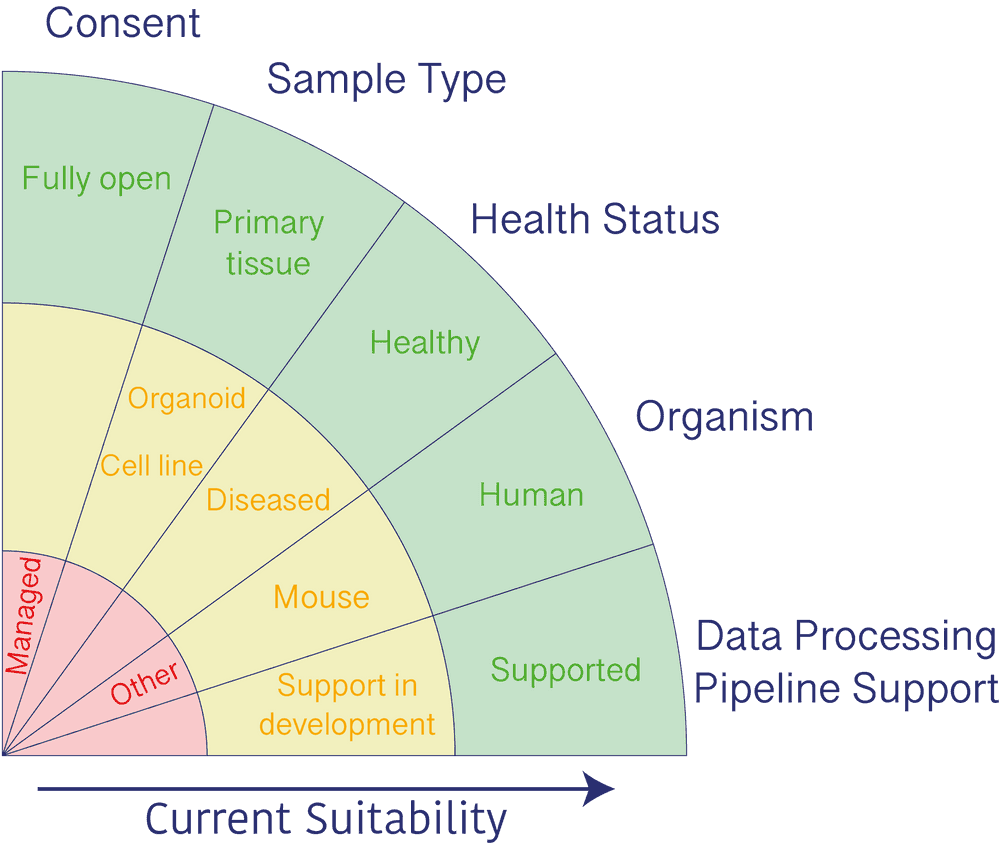
Submitting data to HCA
To submit your data with the metadata in spreadsheet form check out the spreadsheet guide
If you prefer code checkout the guide to submitting data programmatically and the metadata standards.
You can email the HCA data portal’s data wrangling team with any questions: wrangler-team@data.humancellatlas.org
HCA also has an analysis tools registry for any tools that you produce for analyzing this data and to which you can contribute.
6.2.1.4 GenBank
- Stores whole genome sequencing data and assembled genomes
6.2.1.5 ENA (European Nucleotide Archive)
- Sequencing data
6.2.2 Imaging Data
‘Global Bioimaging’ (Swedlow et al. 2021 [cito:agreesWith] [cito:citesAsAuthority] [cito:citesAsRecommendedReading] [cito:discusses]) is a group founded to: “to disseminate best practices, develop common imaging and data standards that promote data sharing”. They describe a distinction between ‘image data archives/repositories’ and ‘added-value databases (AVDBs)’. The Image Data Resource (IDR) is an added-value database whereas the Bioimage Archive is, as the name suggests, an archive/repository. (see the dedicated sections on these below).
The envisioned workflow for image generation, storage and sharing is outlined at a high level in three steps:
- Local Data Storage (pre-publication)
- Archive/Repository
- Added-value Database
This might for example go:
- a local OMERO instance
- Bioimage Archive
- IDR
Only data with sufficient value to be archived should make it from local data storage into public data repositories. If it is data that underpins a published result then it should be archived. Once in a public archive that data should ideally only be referenced by AVDBs rather than replicated to them to avoid unnecessary duplication. Work is underway at the Bioimage Archive to implement APIs which should make it easier to submit data to the archive directly from a local OMERO instance. This should also make further curation/annotation efforts in the AVDBs like IDR & EMPIAR easier.
When publishing results based on image data their value and the effectiveness of the communication of your results can be increased by following community developed standards such as those described here: (Schmied et al. 2023 [cito:citesAsRecommendedReading]). These recomendations come with simple checklists to follow for publishing images and image analysis workflows.

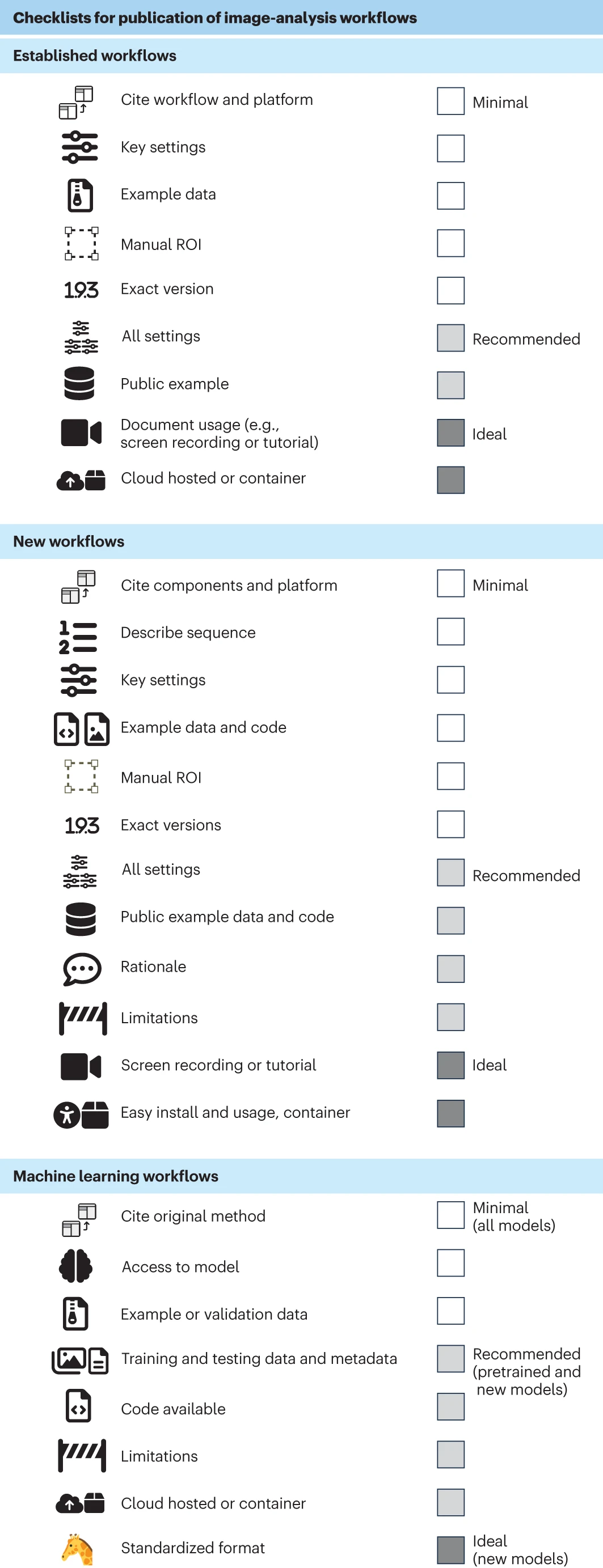
When depositing images in repositories such as the BIA then the REMBI (Recommended Metadata for Biological Images) standard provides excellent guidance for making image data FAIR (Sarkans et al. 2021 [cito:citesAsRecommendedReading]).
6.2.2.1 IDR (Image Data Resource)
- An added value database of high quality well annotated bio-image data of cells and tissues
- Quite extensive manual curation, currently only accepting ‘reference’ collections with high potential for re-use.
- IDR is an instance of OMERO, thus managing your metadata in a local instance of OMERO should make it easier to release result here.
6.2.2.2 Bioimage Archive (EBI)
- An Archival repository of biological images
- Broad scope, any scale or modality
- Associated with a publication or resource of general interest.
6.2.2.3 EMPIAR (Electron Microscopy Public Image Archive)
- An Added value database for Electron microscopy data
6.2.3 Protocols
Useful article in Nature about writing reproducible lab protocols(Baker 2021 [cito:citesAsRecommendedReading]).
This European Commission Joint Research Centre Report: Promoting reusable and open methods and protocols (PRO-MaP): recommendations to improve methodological clarity in life sciences publications (Batista Leite et al. 2024)
Has these key recomendations for researchers:
- Document, share and follow protocols within your research group
- Follow study design and reporting guidelines when designing and conducting your studies and reporting results
- Describe methods in enough detail to allow others to reproduce the experiments
- Ensure availability of methods and materials reported in papers and publications
- Support a culture that rewards and incentivises method development and protocol sharing
Another point to highlight from this report is the importance of the responsible use of methodological short-cut citations:
Researchers use a shortcut citation when they cite a resource that used the method instead of fully describing the method themselves. Researchers should ensure that cited resources are accessible and contain a detailed description of the methods that the citing authors used.
6.2.3.1 Protocols.io
- Publish details of your laboratory protocols. Step-by-step procedures optionally supplemented them with images and other media as a supplement to the textual descriptions of the methods. (Unfortunately protocol.io is proprietary platform operated by a private company (Springer Nature) not a publicly owned archive or open source tool but I’m not aware of any good alternatives at the moment.)
6.2.3.2 JOVE
- (Journal of Visualized Experiments) Publish videos of how you perform your experimental work. This makes it easier share intricate experimental details not readily captured in text.
6.2.4 Code & Computational Environments
Checkout the Research Software Sharing, Publication & Distribution Checklists that I have been developing as a guide to the steps of sharing, publishing and distributing different kinds of research software outputs.
6.2.4.1 Software packages
In language specific package repositories
R:
bioconductor, CRAN (Comprehensive R Archive Network), both have review processes for submitting packages to their repositories.Python:
PyPi, anyone can upload
Software publications
If you have written a piece of open source software as a part of your research that stands alone as a substantial scientific output the you might want to turn it into an academic publication with peer review. These slightly alternative journals facilitate that.
6.2.4.2 Bioinformatic analysis pipelines
If you have constructed a robust bioinformatic analysis pipeline that does the sort of data processing that other might want to do as well, then as long as you have used the appropriate tools to build your pipeline there are options to share them with a wider community of researchers.
WorkflowHub for any type of workflow
targetopia for R {targets} pipelines (via rOpenSci) - more focused on composable components of pipelines that can be connected together to perform certain types of analysis that necessarily complete pipelines
6.2.4.3 Scripts, Notebooks and project specific workflows can be shared as git repositories.
on git Hosting platforms: Renku Section 4.8, Gitlab, Github
You can generate DOI’s for your projects with Zenodo
NoteHDBI Zenodo URLsCollection URL:
https://zenodo.org/communities/hdbi/Above address links directly to your community collection.
Upload URL:
https://zenodo.org/deposit/new?c=hdbiAbove address will automatically ensure people who use it will have their record added to your community collection.
Curation URL:
https://zenodo.org/communities/hdbi/curate/Above address links to your private curation URL. You will find all uploads pending your curation.
Harvesting URL:
https://zenodo.org/oai2d?verb=ListRecords&set=user-hdbi&metadataPrefix=oai_dcAbove address links to a OAI-PMH feed, which can be used by other digital repositories to harvest this community.
Docker images - pre-built reproducible computational environments
6.2.5 Biological Materials access / Sharing
6.2.5.1 HDBR (Human Developmental Biology Resource)
- “[HDBR] is organised from two sites: the Institute of Genetic Medicine, Newcastle, and the Institute of Child Health, London. The HDBR is an ongoing collection of human embryonic and fetal material ranging from 3 to 20 weeks of development.”
- See also the HDBR Atlas: “a digital atlas comprising 3D reconstructions from Carnegie Stage 12 to 23, generated using Optical Projection Tomography (OPT), and annotations of the 3D models linked to an anatomical database”
6.2.6 Spatial transcriptomics
A consensus has yet to emerge in this area and different technologies have different underlying datatypes, some use sequencing and some are more array like.
The SpatialData format is emerging as an open format for processed spatial data. It is most similar to that of other image data as it is built on zarr data structures and has been developed in coordination with the OME-NGFF efforts. It is possible therefore that BIA might take submissions in this form, one could consider the expression matrices as rather extensive image metadata.
The Haniffa lab’s webatlas is a good tool for viewing this data especially if it is integrated with single cell transcriptomics.
6.2.7 Flow Cytometry
flowrepository the International Society for Advancement of Cytometry (ISAC) FCS File Repository
- Data deposited in the flow repository should meet the MIFlowCyt (minimum information about a Flow Cytometry Experiment) standard (Lee et al. 2008 [cito:citesAsAuthority]), This paper Guide to preparing data that meets the MIFlowCyt standard (Spidlen, Breuer, and Brinkman 2012 [cito:citesAsRecommendedReading]).
6.2.8 Proteomics
PRIDE is the primary repository for proteomics data. To submit data to them you need an account and to download their Java based submission tool, the process is well documented on their website.
6.2.9 None of the above
If your data does not fit into any of the above categories and you can’t find a public repository that will host it for you then there are number of generalist repositories like Zenodo see this Zenodo publication on choosing a generalist repository, OSF, & Dyrad
If for some reason even these generalist repositories don’t work for you you might consider hosting your own instance of dataverse, DataHub or iRODS which are software projects that provide tooling for managing your own data repository.
6.3 Integrated publishing - a possible future
Data, analysis, prose, collaboration, pre-print, review and publication in one place with literate programming and single source publishing
You begin your project on an instance of a platform like Renku (section Section 4.8), Start by uploading your raw data to a domain specific data repository. You get a DOI or accession for your dataset. You import this into your project. You perform your computational analyses in the reproducible computational environment. Potentially documenting your analysis as a workflow that could be used by others with a pipeline management tool. You write your manuscript in a literate programming format like Quarto. You work with your collaborators on the manuscript using a git hosting tool like gitlab where you raise and discuss issues, and share revised versions. You generate your statistics and graphics for inclusion in the manuscript with code from your data in a reproducible computational environment. You publish a pre-print by making use of a static site generator like the one built into gitlab and simply setting the project to public. You tag this version 0.0.0 and associated it with a DOI from zenodo. To manage reviews of your work you make use of gitlab issues in a manner similar to the review processes of JOSS, rOpenSci and f1000 but potentially independent of a particular publication venue through community peer review projects like Peer Community In (PCI) & Review Commons. This approach permits author led updates, errata, & corrections whilst preserving a version of record (Kane and Amin 2023 [cito:agreesWith]). Once Reviewed and published you have the 1.0.0 version of your manuscript, for future minor corrections you increment the patch version 1.0.1 and your change-log reflects that you fixed a typo. If you add a new dataset or fix an error that changes an outcome you increment the minor version number. If the journal updates the version of record you increment the major version number.
In this Fashion the complete history of the project is documented start to finish and you never had to change medium from scripts to manuscripts in word processors to emailing pdfs, to publisher websites etc. Review is handled with the same set of tools as was your internal collaboration with co-authors. pre-print publication is creating a version tag and setting the repo to public. Anyone can pick up your project in it’s entirety and play around with their own variants of your analysis at the click of a button (specifically the ‘fork’ button).